Covalent Labelling Estimation And Normalization (CLEAN) Package
Validation
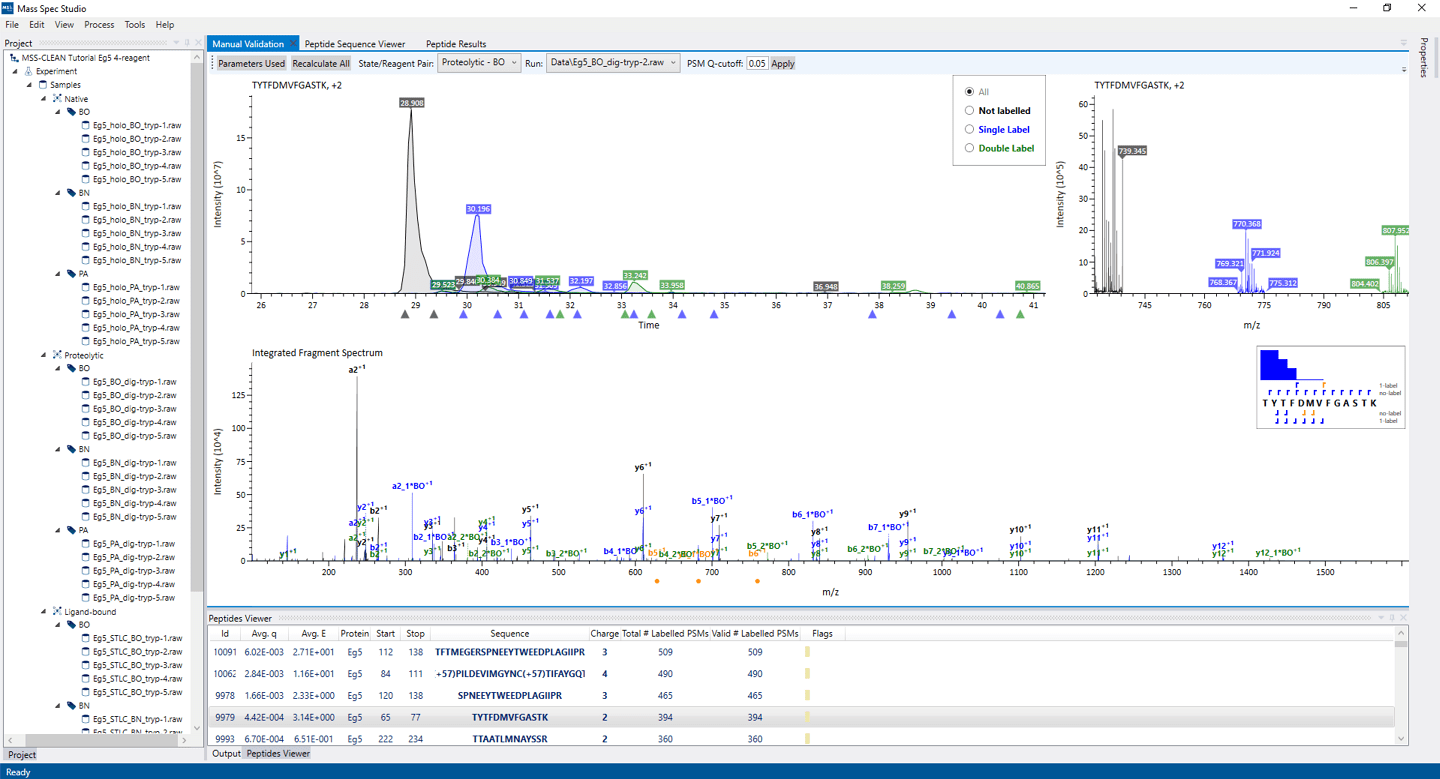
Covalent labeling data is cleanly displayed in a hierarchy based on protein state, reagent and replicate. The validation view allows the user to modify automatic peak selection and to verify correct fragment identification.
Automated processing is enabled via the “Reprocess All” button. Automated peak area selections can be adjusted by click and dragging left-to-right to select an area and right-to-left to deselect. Peak areas need not be continuous.
Inset sequence and column graph shows the identified fragments and relative residue labeling yields. Orange fragments correspond to intensity-imputed fragments. For more information please see our publication linked below.
Visualization
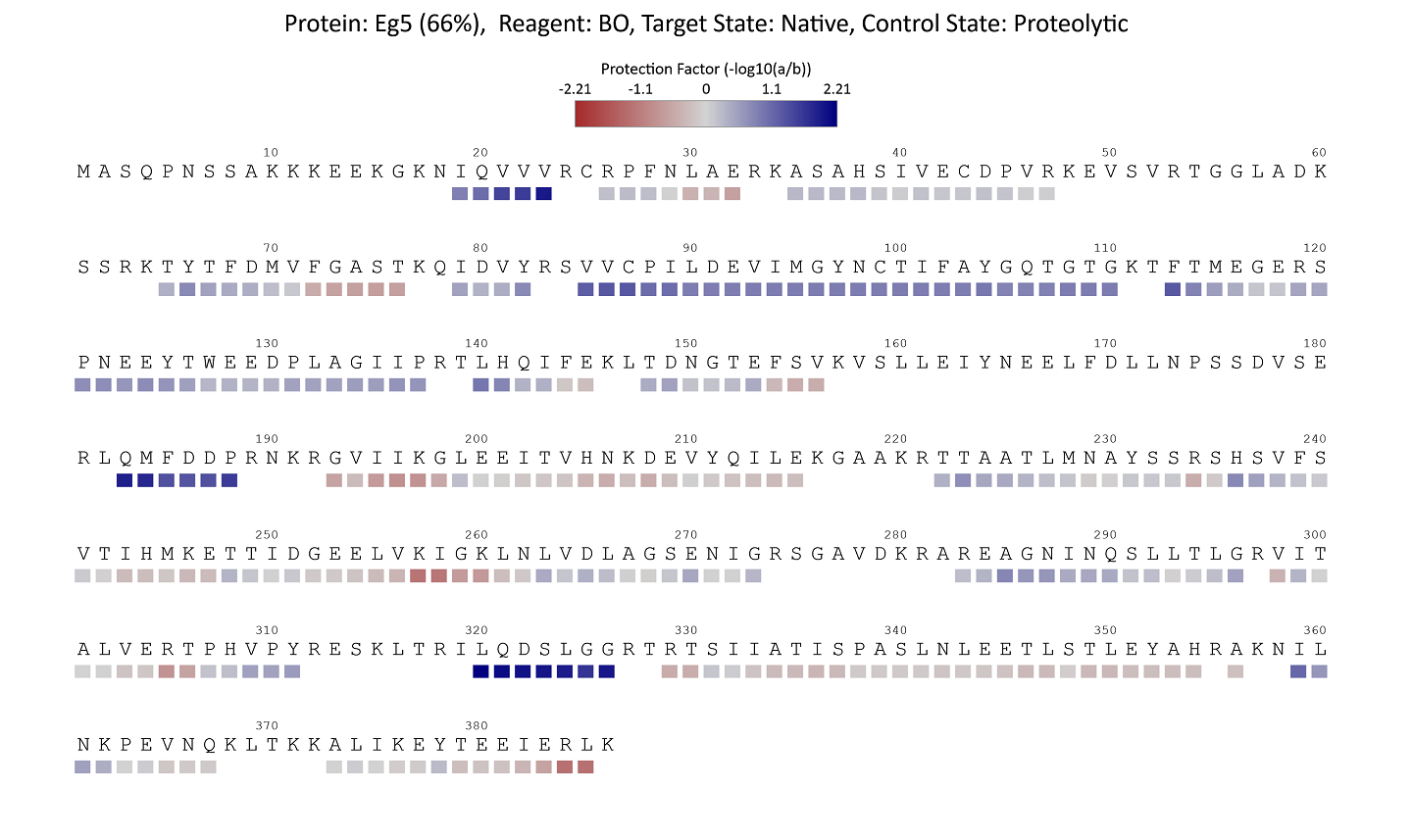
Labeling yields can be previewed on the protein sequence(s) by peptide or residue. Yields may be displayed by absolute or relative yields with respect to a control. Maps can be exported by right-clicking and selecting “Save Image”.
Export
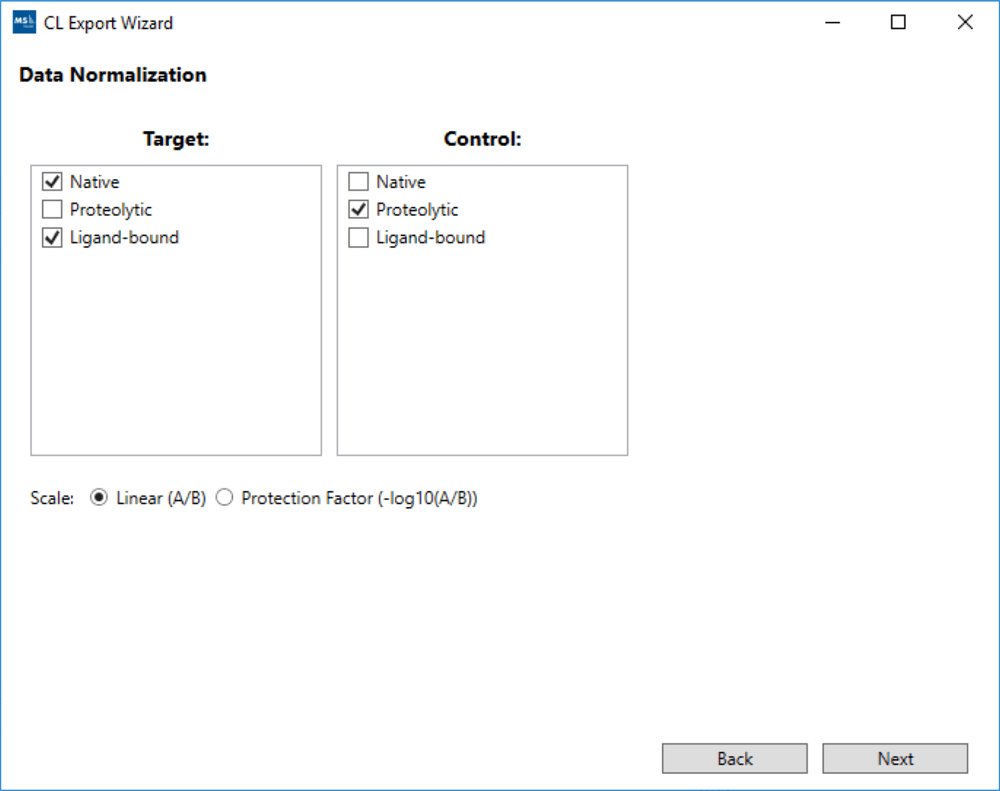
Processed data can be exported in a number of CSV files including normalized yields by residue and a text file ready for mapping to a protein structure via the “Render by Attribute” functionality in UCSF Chimera.
Reference
[Link]: Ziemianowicz DS, Sarpe V, Schriemer DC. 2019. Quantitative Analysis of Protein Covalent Labeling Mass Spectrometry Data in the Mass Spec Studio. Analytical Chemistry.