Rapid development of new components and plug-ins
Experiment Types
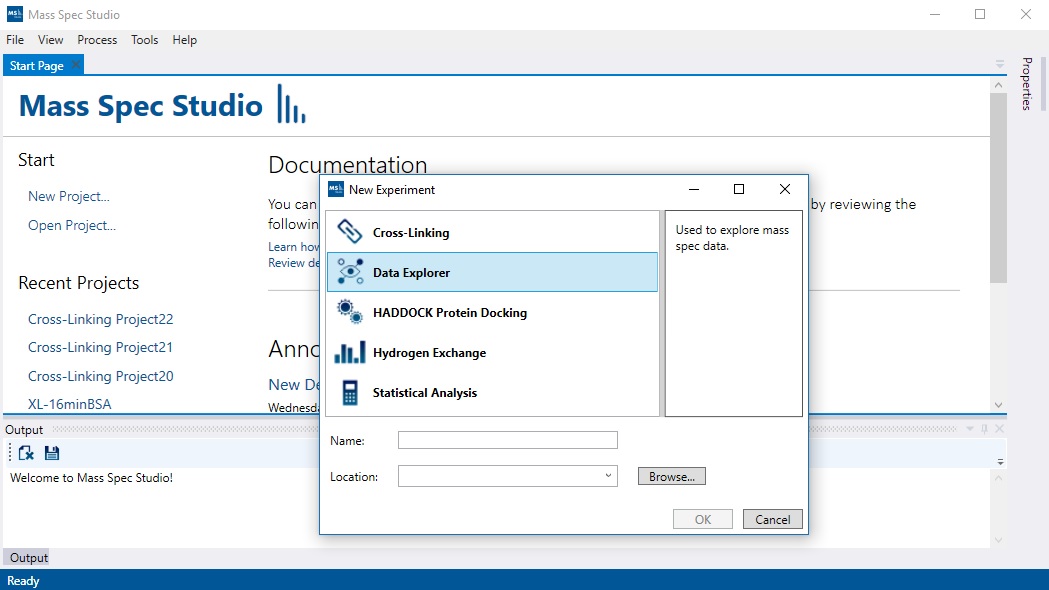
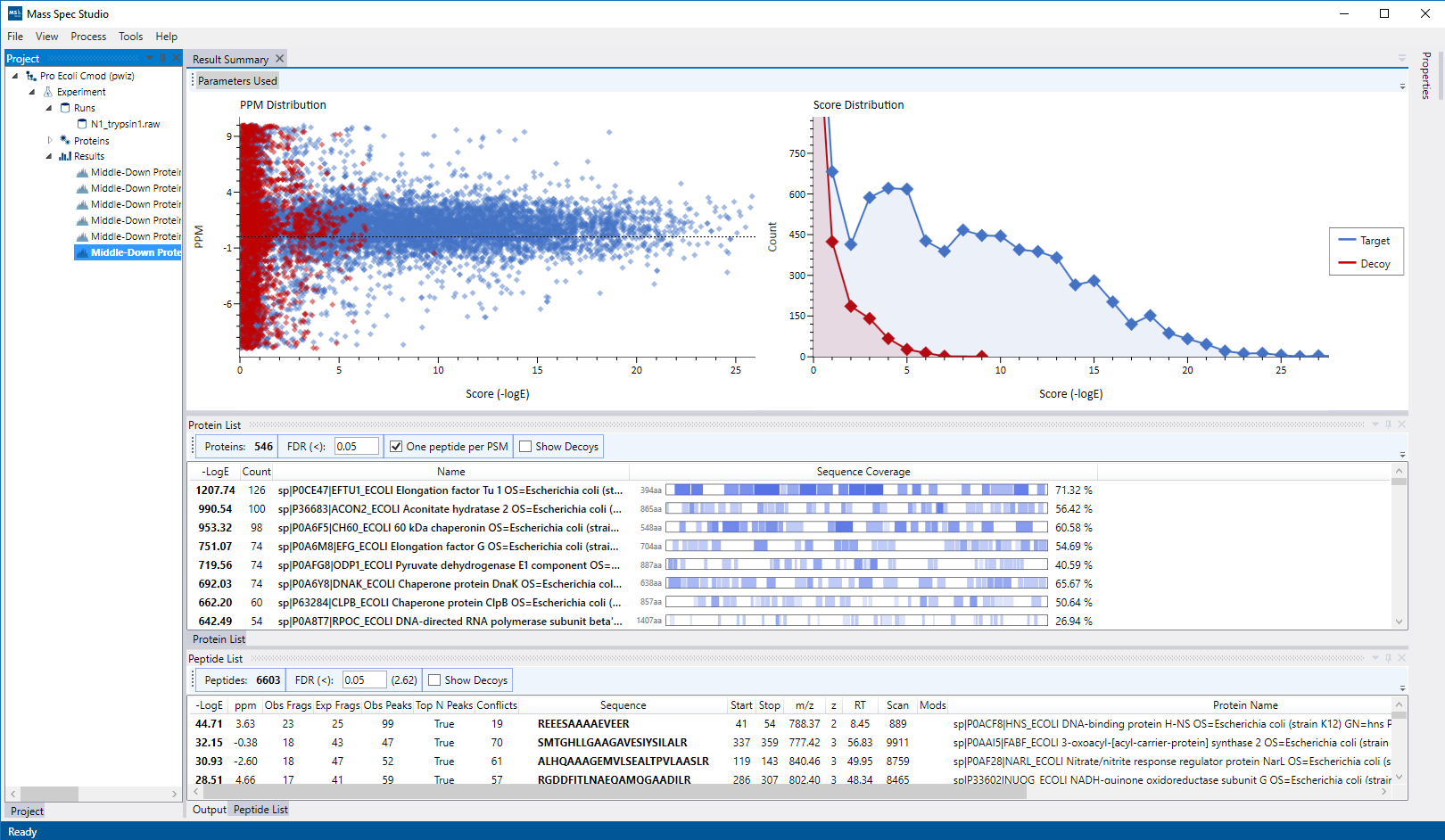
Experiment types serve as ‘entry points’ to a given mass spectrometry project and describe the structure, workflow and requirements of one type of mass spec data analysis activity. Mass Spec Studio allows multiple project types to be installed. This allows users of the application to process different types of data set all from within Mass Spec Studio.
Data Providers
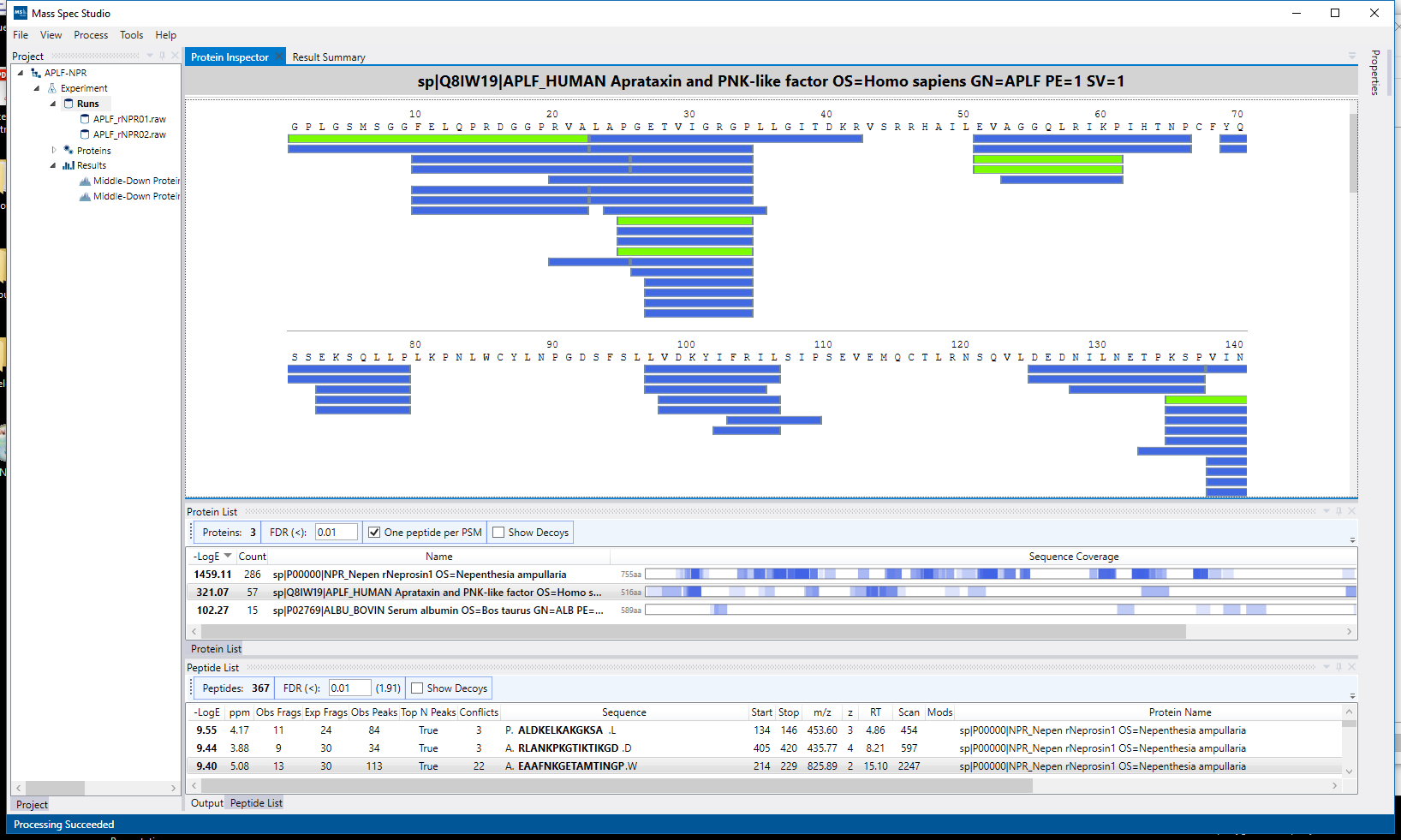
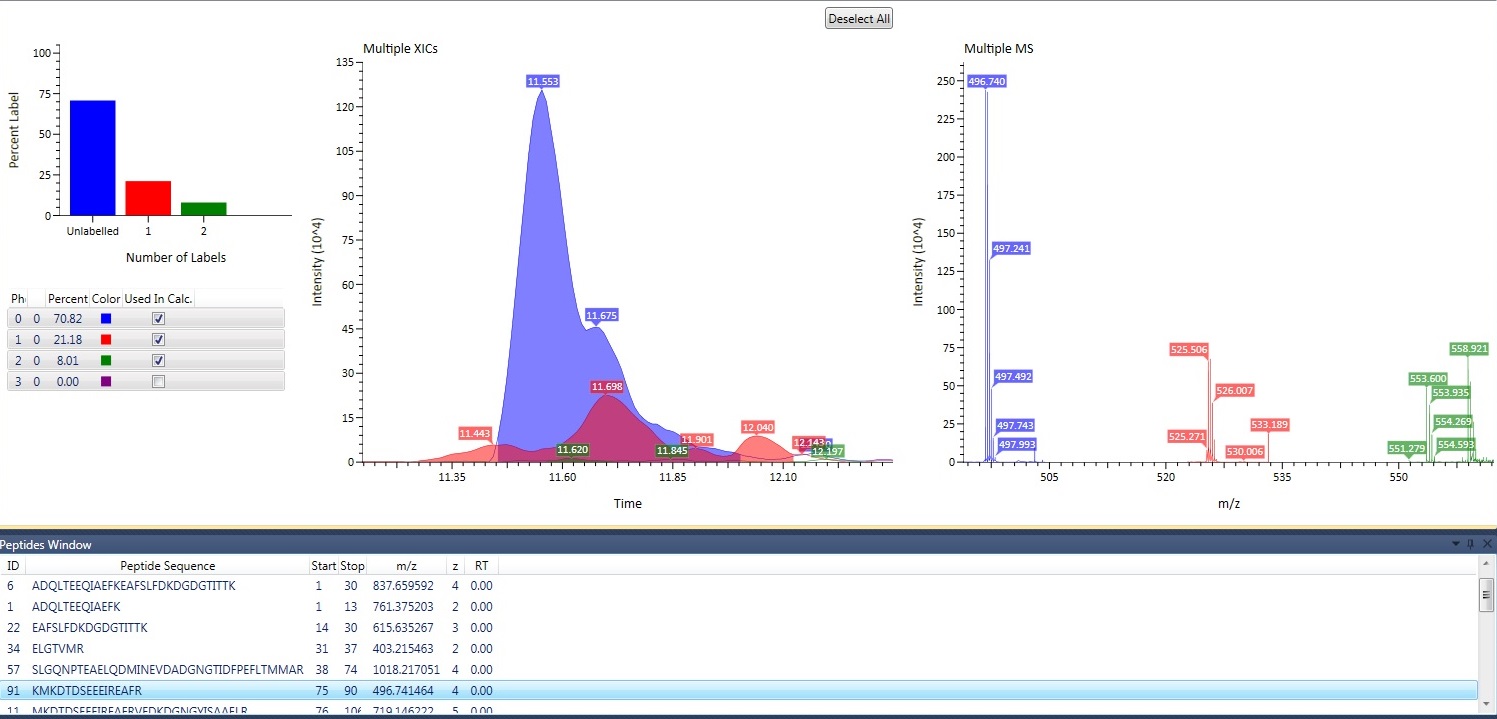
Data providers act as generic data repositories for any mass spectrometry application. Once a data provider is loaded into the framework, any existing experiments, modules or processing components can access and utilize the data regardless of its type or original raw format. Implemented data providers act as wrappers for anything ranging from a simple mzxml reader to a vendor agnostic library like ProteoWizard to even a database hosted on a remote server.
Processing Components
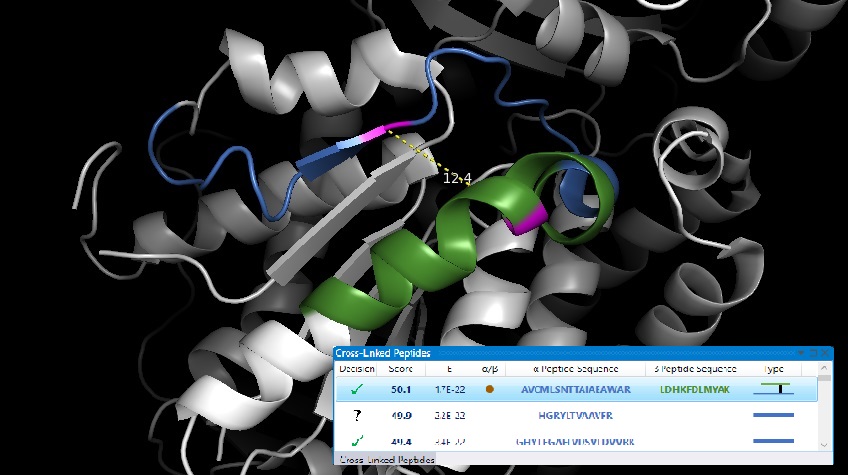
Processing components consist of processing steps and collections of processing steps, namely processing routines. Processing routines act on a project scale, and define customizable processing workflows consisting of multiple processing steps. Processing steps are specialized functions which interact with the underlying data.
UI Modules
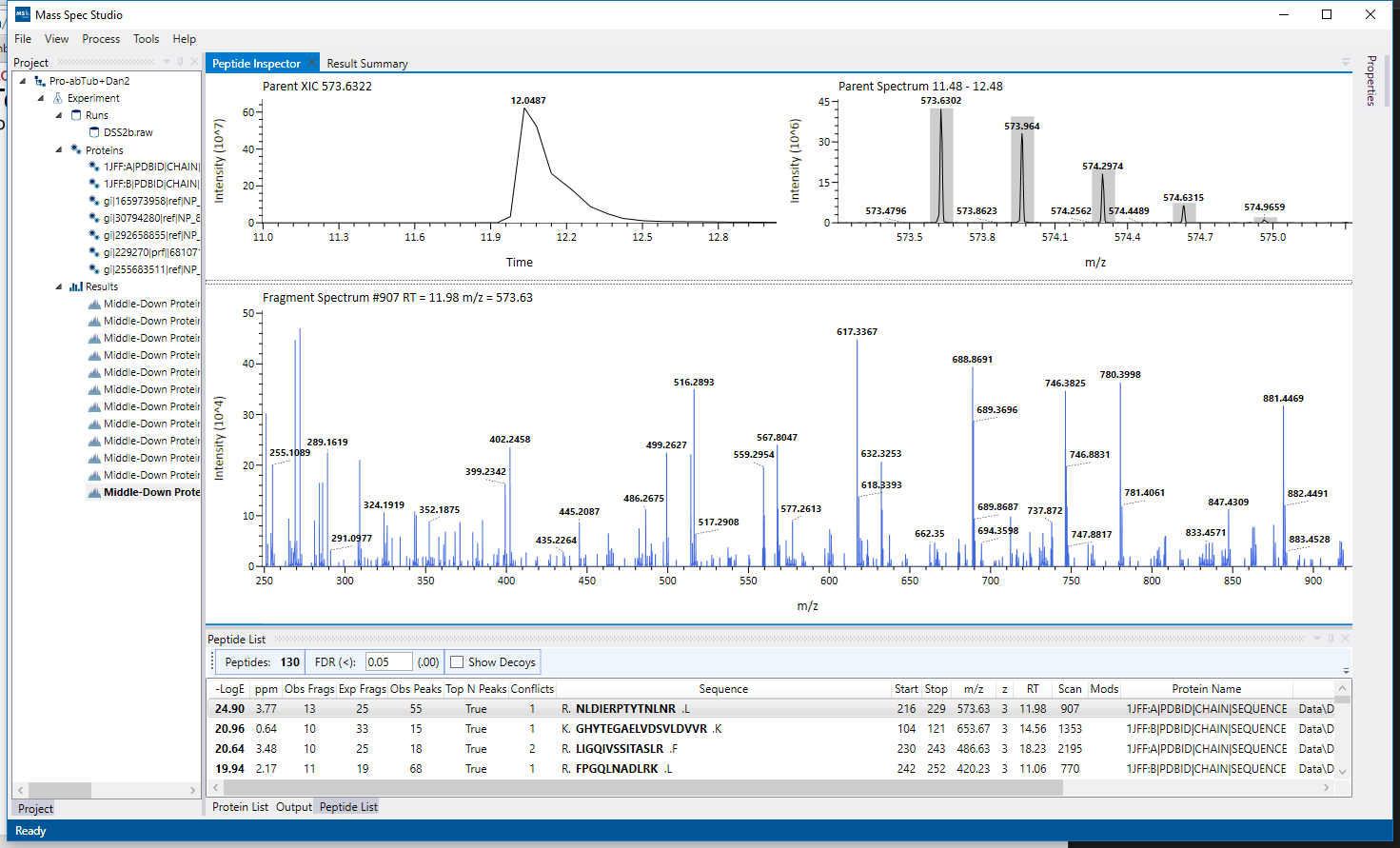
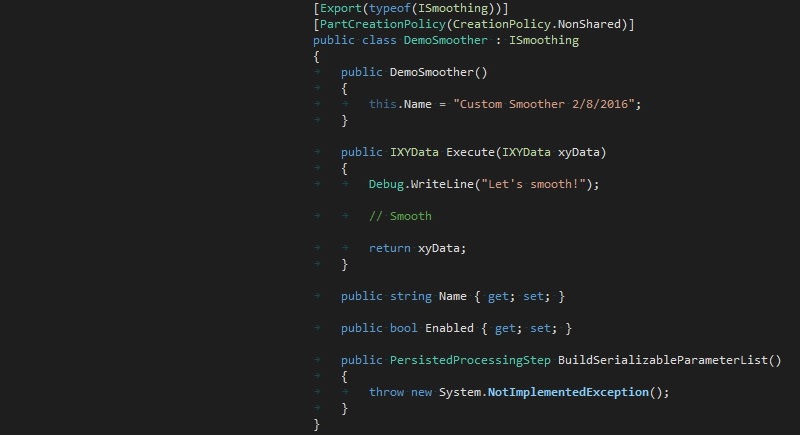
UI Modules are interactive elements (such as views, windows, buttons) which plug into the main application shell. Components which define UI Modules consist of any number of views which are automatically integrated into pre-defined regions of the main application window.