IMProv - Integrative Modeling Platform
Integrative structural biology requires the aggregation of multiple distinct types of data to generate models that satisfy all inputs. Here, we present IMProv, an app in the Mass Spec Studio that seamlessly combines XL–MS data with other structural data, such as cryo-EM densities and crystallographic structures, for integrative structure modeling using IMP (www.integrativemodeling.org).
Configure an IMP run
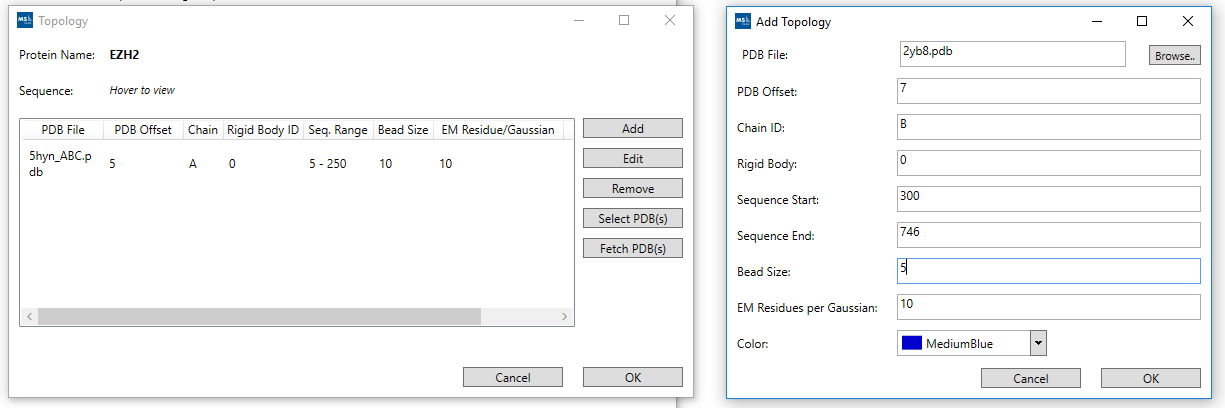
The app contains a wizard to upload protein sequences, atomic structures, all available modeling data, and the necessary graphical interfaces for setting the protein topology files and the configuration of IMP. The output is a set of scripts generated from basic templates. These are combined with IMP to initiate a modeling run.
Containers for running IMP
The scripts and data are combined with a container that includes IMP and the environment for running it. The template script for the IMP Python Modeling Interface (PMI v2) supports cryo-EM and XL-MS data, partial X-ray structures or homologies, as well as basic physical restraints, such as sequence connectivity and excluded volume. We have developed deployment bundles for Amazon Web Services, personal computers and we maintain an installation on Digital Research Alliance of Canada (Niagra cluster). The associated tutorials describe in detail how to take advantage of these resources.
To get started, follow the tutorials in the sidebar.